|
Compiling, running and analysing chains
This file is for the current release, v1.5 (June 2010).
If you are looking
for older versions' read-me files, see the version history
page.
Jump to:
Efficiency scaling
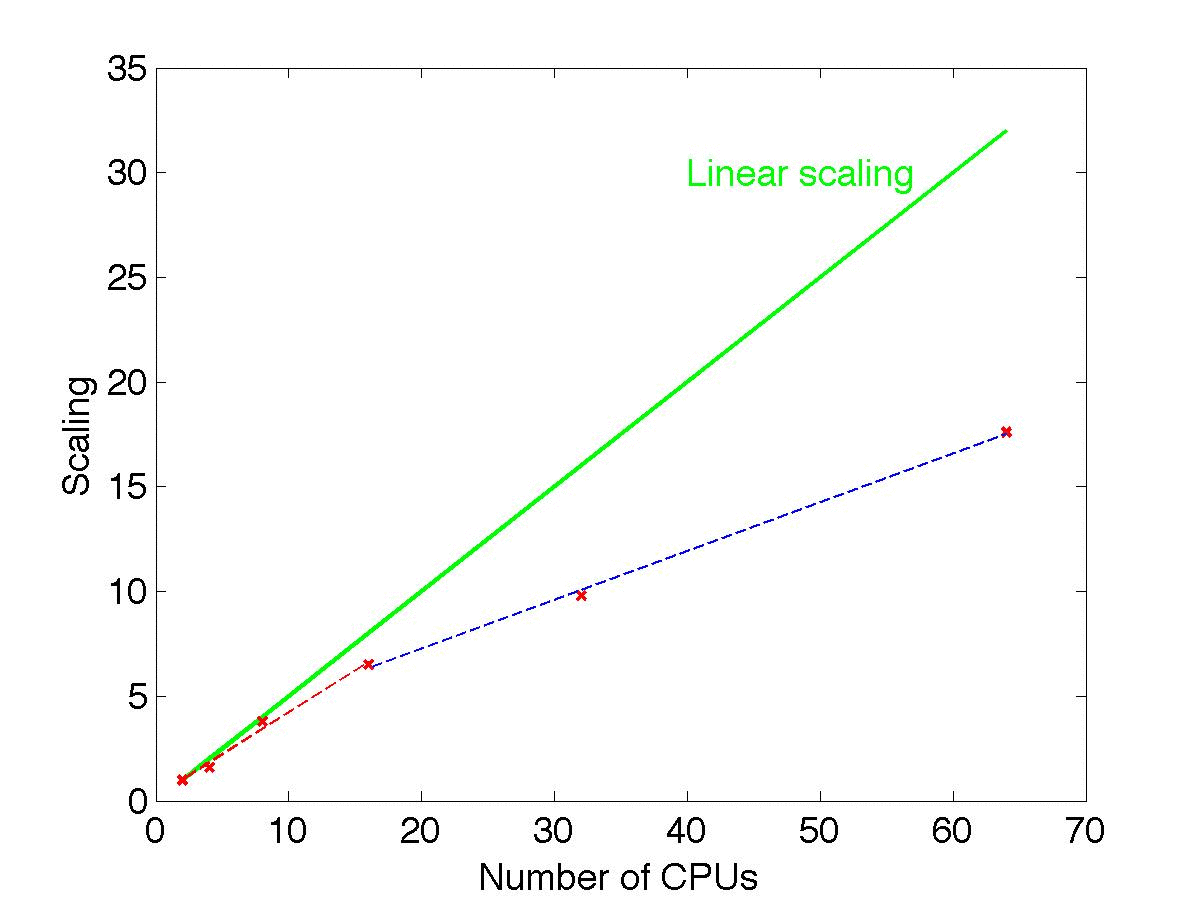 The graph and the table below show the scaling of the performance of SuperBayeS with the number of CPUs used. All runs have been performed on 2.8 GHz, 1600 MHz FSB Intel processors using this ini file (log prior, all data sets included and scanning over 8 parameters, including 4 CMSSM and 4 nuisance parameters).
The graph and the table below show the scaling of the performance of SuperBayeS with the number of CPUs used. All runs have been performed on 2.8 GHz, 1600 MHz FSB Intel processors using this ini file (log prior, all data sets included and scanning over 8 parameters, including 4 CMSSM and 4 nuisance parameters).
Each run took about 4.5 times 10E5 likelihood evaluations to gather around 50'000 samples.
From the point of view of efficiency (in terms of samples obtained per hour and per CPU) the best choice is to run with 8 CPUs. However, notice that approximate linear scaling for the inverse total time required holds up until 16 CPUs (red dashed regression line in the graph, which has slope 0.4, while perfect linear scaling would have slope 0.5, green solid line), and the efficiency gain is only moderately reduced as the number of CPUs is further increased above 16 (the blue dashed regression, with slope 0.25).
| CPUs |
Run time(hrs) |
Samples/hour |
Samples/hour/CPU |
Scaling |
| 2 |
176.2 |
284 |
142 |
1.0 |
4
| 117.7 |
444 |
111 |
1.6 |
8
| 46.1 |
1083 |
135 |
3.8 |
16
| 27.1 |
1873 |
117 |
6.5 |
32
| 17.9 |
2779 |
87 |
9.8 |
64
| 10.0 |
4952 |
77 |
17.6 |
Compiling SuperBayes
The code can be compiled to run on one CPU only or as
an MPI code to run in parallel on an MPI cluster.
In source/Makefile, turn the
FC flag to mpif90
for MPI support or to ifort
to work in single mode.
In MPI mode, if running MCMC the number of chains is automatically set
equal to the number of processors used in the run.
Each chain then produces output files with an id tag "_i",
with i=1,...,n (n being the number of processors). In Grid
Scan mode, the MPI mode distributes points on the grid
across nodes. With MultiNest, MPI mode
switches on the faster MultiNest parallel computation on
the given number of nodes (recommended n=10).
If MPI is off, in MCMC only one chain is produced while
MultiNest runs in serial mode.
From the source directory, the command
make cleanall
cleans all the compiled files and executables.
For a MPI to single processor mode transition use
the command
make clean and reset the
FC flag to ifort
in the Makefile as it was mentioned above.
The command
make all
then recompiles the whole package building
static libraries for each of the codes
included into the package (notice that MicrOMEGAs also
uses dynamical libraries). Then two binary files are produced:
superbayes (which is to be used for runs in MCMC mode, postprocessing and grid scanning) and
superbayes_multinest (to be used for MultiNest mode runs).
The command
make superbayes (superbayes_multinest)
only recompiles files (for the corresponding mode) in the source
directory. Use
make getplots
to compile the getplots
routine (for chain analysis and plotting).
Testing SuperBayes
For testing purposes the testing.90 file is
provided. The command
make tester
will compile it.
The run is made from the command line with
the command
tester
The parameters for running the tester are hardcoded in the
source\tester.f90 file, and are the same as
described below. Setting debug=.true. will write
the full output with detailed info about the point
being considered to the file spectrum.out.
By default a file called ifort.* containing
the formatted output is created too.
The tester works in single-point mode if
test_chain = .false., otherwise it can read in a
list of points from an existing chain
(filename of the chain file hardcoded in tester.f90),
which can be useful for testing purpose.
If your installation has been successfull, the output of
running tester should match the content of the file
tester.output in the main directory.
Running SuperBayeS
If MPI is turned off, Superbayes is invoked in
single-processor mode from the command line with the
command
superbayes (superbayes_multinest) SampleIniFile.ini
If you try e.g. to run superbayes in MultiNest mode, you will get an error message (and viceversa).
The corresponding MPI command depends on the
configuration of your machine. The SampleIniFile.ini
file contains all parameters for the run.
Currently, only the Constrained MSSM is supported,
but the package is easily customizable
to extend the scan to the general MSSM if required.
The syntax of the .ini file is mutuated from the
CosmoMC package,
and the meaning of the parameters is
explained here
.
SuperBayeS can be run in MCMC mode (using
Metropolis-Hastings), in grid-scan mode (which returns
the likelihood on a multi-dimensional grid at pre-defined
spacings in parameter space) or (recommended option) in
MultiNest (Nested Sampling) mode (use the superbayes_multinest binary).
See running
options for details.
(top)
Analysing the chains and plotting
SuperBayeS comes with GetPlots, a routine for
analysing the MCMC and MultiNest outputs and plotting the
results in 1, 2 and 3 dimensions. This is based on GetDist,
from the CosmoMC package - refer to
CosmoMC website
for futher details. The current version has many extensions
and new facilities that are described in detail
here
GetPlots is invoked with the command
(from SuperBayeS root directory)
getplots GetPlotsSample.ini
Output files are stored in subdirectories of the folder
output_files folder (any pre-existing files
are overwritten).
Those files contain the statistical information about the
run and the matlab and SuperMongo (SM)
files needed to produce plots (see
here for details).
Data files needed by matlab and SM for the plots are stored in
subdirectories of the folder plot_data
(it should usually not be necessary to edit or otherwise
change these files).
To generate .ps plots, go to the
output_files/rootname directory and
call SM (for 1D plots) with the command
sm < rootname_1D.sm
or matlab via the command
matlab < rootname_1D.m or matlab < rootname_2D.m or matlab
< rootname_3D.m
Details of the format of the ensuing plots
(line colours, thickness, labels, colormaps, etc) can be
custom-edited in the source files
source\matlab.f90 (matlab plots) and
source\smplots.f90
(SM plots).
Analysis and plotting options are
explained here. Interactive plotting with SuperEGO is described
here.
The list of output files and their meaning is
explained here.
(top)
Running options (indirect detection quantities
options are listed separately here.
MultiNest specific running options are
here )
-
file_root: the and location (and prefix) of the output
files produced. The list of file produced depends on
the running mode, see here .
When postprocessing,
this becomes the name of the input chains.
If restart = T,
the job restarts from the last line of the chains or
from the last sampling step for MultiNest. If
MCMC/MultiNest chains with the same name already exist,
they are overwritten. In MCMC mode only, to prevent the
overwriting from happening, set the variable
FailSafeOn = .true. (hardcoded)
in source/utils.F90
-
out_root: the name of the output
files produced when postprocessing (otherwise
ignored).
-
restart_and_continue: set it to F
to start a new job, set it to T to continue from
where a previous chain stopped. In MultiNest mode,
don't change nlive
option while restarting a run otherwise the program
would abort.
-
action: determines how the scanning
is performed:
-
action = 0 to do
MCMC (currently only Metropolis-Hastings is
supported). Set the lambda parameter to a value > 0 to activate the bank sampling mode. If lambda > 0 it is assumed that a bank samples file exists with name file_root_bank.txt .
-
action = 1 to post-process an
existing set
of chains (useful for computing new variables, or
doing a rough posterior adjustment for new data or
new priors without re-running the whole chain, or
for computing the indirect detection quantities
corresponding to the output of a MCMC or MultiNest run)
action = 4 to compute the likelihood
on a
fixed-grid in parameter space
(if MPI is enabled, each chain covers one part of
the grid). When run in grid-scanning mode, only
physically admissible points (e.g., EWSB achieved,
no tachyonic masses, etc) are saved.
-
action = 5 for MultiNest
(recommended)
-
When post-processing, redo_like = T
will recompute the likelihood from the saved values
of the variables in the chains, without recomputing
the theory. This is useful if only the data have
changed, but you must have saved all of the relevant
variables in the chains, as observables are not
recomputed. redo_theory = T will
recompute the observables, as well (useful if the
theoretical predictions have changed).
redo_change_like_only = T
will just change the likelihood (i.e., multiplicity of the samples is not affected. NB: this is not recommended except for testing purposes).
-
skip_lines: number of samples
that are not saved at the beginning of the MCMC run
(burn-in period).
You might as well save them and remove them later
when analysing the chains.
-
Use_MICRO: set it to T
uses MicrOMEGAs for computing the
relic density of dark matter abundance.
Otherwise DarkSusy is used instead.
Notice that currently dark matter direct and indirect
detection quantities are computed by DarkSusy
anyway.
-
compute_xxx: those flags determine
which quantities are computed and saved in the .txt
files:
-
compute_DM: set it to T to compute
the relic dark matter abundance.
CDM_purely_LSP = T assumes the LSP
is the dark matter, otherwise the dark matter is
made up of LSP plus another component and hence the
WMAP3 observations are only an upper bound
(the latter mode is currently untested).
-
compute_Direct_Detection : set it
to T to compute direct detection cross sections.
-
compute_Indirect_Detection: set it
to T to compute indirect detection quantities.
-
compute_Collider_Predictions:
set it to T to compute collider-related quantities
(masses, etc).
-
compute_BD: set it to T to compute
B decay predictions.
-
compute_FH: set it to T to compute
cross sections and branching ratios using FeynHiggs.
Customize the variables you want to save by modifying
the routine ReduceOutput in
source/paramdef.f90 and the corresponding
type, Reduced_Out
-
feedback: controls amount of text
printed on standard output. 0 = none, 1 = some, >2
debug mode.
-
use_xxx: those flags determine
which quantities are used in the computation of the
likelihood (obviously if you want to use them you
have to set the relative compute_xxx
flag to T). All of the data values are found
in the file likedata.f90.
Refer to
our paper for how
the likelihood is computed. Meaning as below.
-
Use_Nuisance: set it to T to use
current constraints for nuisance (SM) parameters.
-
Use_CDM: set it to T to use current
cosmological constraints on dark matter abundance.
-
Use_LEP: set it to T to use
constraints from LEP on sparticle masses and Higgs
mass.
-
Use_Anomalous_Mu: set it to T to
use constraints on the anomalous magnetic moment of
the muon.
-
Use_bsgamma: set it to T to use
constraints on the process B-> s gamma.
-
Use_Bsmumu: set it to T to use
constraints on the process B-> mu mu.
-
Use_Mass_W: set it to T to use
constraints on the W mass.
-
Use_Weak_Mixing_Angle: set it to T
to use constraints on the effective weak mixing angle.
-
Use_Delta_MBs: set it to T to use
constraints on Bs-Bs oscillations.
-
Use_Butaunu: set it to T to use
constraints on the process B-> nu tau.
-
Use_DD: set it to T to use
constraints on direct dark matter detection
(currently not supported).
-
Use_ID: set it to T to use
constraints on indirect dark matter detection methods
(currently not supported).
-
use_data: select from the list to
use current data (Jan 2010) or constraints from future
observations (edit your future data
in source/likedata.f90).
-
propose_matrix: set it to the name
of the file containing the covariance matrix from
previous runs. Used to adjust the proposal width in
the new run (MCMC only).
-
redo_likeoffset: when
postprocessing it might be useful to put an offset
to the loglike if there is a large change to it with
new data to get sensible weights.
-
samples: number of samples to
obtain per chain (MCMC only). All accepted samples are counted
(after burn-in). If in grid-mode, this sets a limit
to the maximum grid points per chain that will be
allowed - increase it as needed.
-
temperature: temperature of the
MCMC (1 by default). Increase to explore the tails,
jump more easily to disconnected regions of parameter
space, etc (must be matched by the cooling factor
when analysing the chains).
-
rand_seed: if blank this is set
from system clock.
-
use_log: whether to use a log scale
(set it to T) or a linear scale (set it to F) for the
gaugino and scalar mass parameters.
-
param_xxx: parameters over which to
do the scanning. For MCMC and MultiNest, flat
priors are taken on this set of parameters.
The meaning of the 5 real numbers is the following:
-
For MCMC: start_central_val, min_val,
max_val, start_width, propose_width
where start_central_val is the
central value around which the chain is started,
min_val/max_val are the minimum and
maximum values allowed (prior range),
start_width is the standard
deviation around start_central_val from which the
starting point is drawn,
propose_width is the proposal
width for the Metropolis-Hastings step (overriden if
a covariance matrix is present).
-
For grid scanning: ignored, min_val,
max_val, ignored, grid_step
where ignored is irrelevant,
min_val/max_val set the grid's
boundaries and grid_step
gives the step size in that direction.
The grid is split among chains if running in MPI mode.
If the number of grid points per chain exceeds the
number of samples, you will be asked to increase
samples.
-
For MultiNest: ignored, min_val,
max_val, ignored, ignored
where ignored is irrelevant,
min_val/max_val set the ranges of the priors
The number of samples is automatically determined by the tolerance level requested for the evidence value.
(top)
Indirect detection quantities
options
All of the following options are only relevant
if compute_Indirect_Detection is set to true.
We recommend using the post-processing routine to compute
indirect detection quantities from the saved MCMC or MultiNest run rather
than computing them directly in the run.
-
compute_ID_gadiff: set it to T to
compute the differential spectrum of gamma-ray.
-
compute_ID_gacont: set it to T
to compute the gamma-ray flux with continuum energy
spectrum integrated above some threshold energy.
-
compute_ID_gamonoc: set it to T
to compute the monocromatic monochromatic gamma-ray flux
induced by 1-loop annihilationprocesses into a 2-body
final state containing a photon. There are two such final
states: the 2 photon final state and the final
state with a photon and a Z boson.
-
compute_ID_antiprot: set it to T
to compute the differential spectrum of antiprotons.
-
compute_ID_antideut: set it to T
to compute the differential spectrum of antideutrons.
-
compute_ID_posit: set it to T
to compute the rates of positrons.
-
compute_ID_positfrac: set it to T
to compute the positron fraction.
-
compute_ID_muonsun: set it to T
to compute the total rates in neutrino telescopes from
the Sun above some threshold energy.
-
compute_ID_muonearth: set it to T
to compute the total rates in neutrino telescopes from
the Earth above some threshold energy.
-
compute_ID_sundiff: set it to T
to compute the differential rates in neutrino telescopes
from the Sun.
-
compute_ID_muonearthdiff: set it to
T to compute the differential rates in neutrino
telescopes from the Earth.
-
compute_ID_eqbsun: set it to
T to compute the capture and annihilation rates
of neutralinos at Sun.
-
compute_ID_eqbea: set it to
T to compute the capture and annihilation rates
of neutralinos at Earth.
-
compute_ID_musuevent: set it to
T to compute the number of events produced at
ICECUBE from neutralinos annihilation to neutrinos
at Sun.
-
compute_ID_muonhalo: set it to T
to compute the total rates in neutrino telescopes from
the halo above some threshold energy.
-
compute_ID_muonhalodiff: set it to T
to compute differential rates in neutrino telescopes from
the halo.
-
compute_ID_sigmav: set it to T
to compute the annihilation cross section sigma v at p=0
for neutralino-neutralino annihilation.
-
compute_ID_efluxes: set it to T
to compute differential fluxes over a range of energies
for each model. It only works in the postprocessing mode.
-
num_hm: sets the number of halo
profiles used.
-
modelxx: sets the specific halo
models you want to use
(see
DarkSusy).
-
pbmodel: sets the antiprotons
and antideutrons propagation model one wants to use
(see
DarkSusy).
-
epmodel: sets the positron diffusion
model (see
DarkSusy).
-
ntmodel: sets the neutrino telescopes
parameters (see
DarkSusy).
-
cospsi0: for gamma-rays and
neutrinos with the chosen halo profile, sets the line
of sight integration factor j in the direction
of observation, which is defined as the direction
which forms an angle psi0 with respect to the direction
of the galactic centre (see the .ini file).
-
delta_gamma: for gamma-rays
if one takes into account the angular
resolution of the detector then delta_gamma is
set is given (in sr) otherwise it is set to 0
(see the .ini file).
-
egam: sets the energy (GeV) for the
differential gamma-ray flux.
-
egath: sets the threshold energy
(GeV) for the gamma-ray flux with continuum energy
spectrum.
-
BF: sets the boost factor
for antimatter fluxes.
-
epb: sets the kinetic energy (GeV)
of the antiprotons for the differential flux of
antiprotons.
-
edb: sets the kinetic energy (GeV)
of the antideutrons for the differential flux of
antideutrons.
-
eep: sets the kinetic energy (GeV)
of the positrons for the differential flux of positrons.
-
eth: sets the energy threshold (GeV)
for neutrino telescopes.
-
thmax: sets the
maximum half-aperture angle (degrees) for
neutrino telescopes.
-
enu: sets the neutrino energy (GeV)
for differential flux of neutrinos.
-
theta: sets the angle from
center of Earth/Sun in degrees for neutrino telescopes.
-
rtype: sets the type of fluxes
(see the .ini file).
-
delta_nt: as delta_gamma for neutrino
fluxes.
-
exposure: sets the exposure time
(yrs) for the computation of events at ICECUBE.
-
ic_config: sets the ICECUBE
string configuration for the computation of events
(see the .ini file).
-
efluxes_i: sets the initial
energy of the fluxes spectrum computation once
the compute_ID_efluxes is on.
-
efluxes_f: sets the final
energy of the fluxes spectrum computation once
the compute_ID_efluxes is on.
-
nbins: sets the number of the
bins to be scanned in the fluxes spectrum computation.
Notice that it is in logaritmic scale.
Running options for MultiNest
-
multimodal: whether to produce separate statistics and samples for each found mode. For problems with several modes with vastly different amplitudes, setting
multimodal to T stops live points from migrataing to dominant modes from weaker modes and therefore allows all the modes to be explored at greater depth. If the problem is inherently
multi-modal but multimodal is set to F, the sampling is still done from all the modes but only the modes contributing significantly to the evidence are explored in any detail.
-
maxmodes: the maximum number of modes expected. This is relevant only if multimodal = T and is used for memory allocation only. If more modes are found than
maxmodes then the program would abort with the error message "ERROR: More modes found than allowed memory. Increase maxmodes in the call to nestrun and run MultiNest again. Aborting".
The user can then resume the sampling by setting maxmodes to a higher value and running SuperBayeS again with restart_and_continue set to T.
-
nCdims: no. of parameters for mode separation. This is relevant only if multimodal = T. Mode separation is done through a clustering algorithm. Mode separation can be
done on all the parameters (in which case nCdims should be set to ndims, the total no. of sampling parameters) & it can also be done on a subset of parameters (in which case nCdims <
ndims) which might be advantageous as clustering is less accurate as the dimensionality increases. If nCdims < ndims then mode separation is done on the first nCdims parameters. For
CMSSM, the recommended value is 2 (with mode separation being done on m_0 and m_{1/2}).
-
ceff: whether to run MultiNest in constant efficiency. If ceff is set to T, then the enlargement factor of the bounding ellipsoids are tuned so that the sampling
efficiency is as close to the target efficiency (set by eff) as possible. This does mean however, that the evidence value may not be accurate.
-
nlive: the total no. of live points. The recommended values are as follows (see 1101.3296 for details):
- For an accurate mapping of the Bayesian posterior, nlive = 4000 .
- For an accurate computation of the Bayesian evidence, nlive = 4000 .
- For an accurate mapping of the profile likelihood, nlive = 20000 .
-
eff: the maximum sampling efficiency. A value greater than 1 means that the MultiNest will sample from a region with volume smaller than the volume enclosed by the
prior volume at any given iteration. The recommended values for parameter estimation & model selection are 2.0 & 1.0 respectively. If running in constant efficiency mode (i.e. when ceff
= T), eff is the target efficiency and its recommended value is 0.1 or lower.
-
tol: defines the stopping criteria. The recommended values are as follows (see 1101.3296 for details):
- For an accurate mapping of the Bayesian posterior, tol = 0.5 .
- For an accurate computation of the Bayesian evidence, tol = 0.5 .
- For an accurate mapping of the profile likelihood, tol = 0.0001 .
(top)
Plotting options for getplots
Plotting options are set in the
GetPlotsSample.ini file to be used with
getplots as follows:
-
file_root: name of chains to be
analyzed (including chains subdirectory). Numbering
of chains set automatically using the
chain_num setting.
-
out_root: name of output files
(if empty, the same as file_root)
-
add_columns: number of new
combinations of variables to add to the anaylsis.
Customize this by writing your own
AddParams routine in
GetPlots.f90
-
smoothing: set to T to use Gaussian
smoothing with a kernel about 3 bins wide. Useful
to reduce jaggedness of 2D contours. Set it to F to
use top-hat bins (no smoothing).
-
chain_num: number of chains to
process. If 0 it assumes one chain and no filename
suffixes. The code automatically looks for MultiNest
output if it does not find an MCMC chain.
-
first_chain: default is 1.
-
exclude_chain: if you want to
exclude one particular chain from the analysis.
-
num_bins: number of bins per
dimension.
-
skip_bin: number of bins to discard
at the edges (use with care).
-
ignore_rows: number of rows to
discard when analysing (burn-in period. Note this
should be used only with MCMC-generated chains.
MultiNest does not do any burn-in and therefore no rows
should be discarded).
-
cool: cooling factor, must match
the temperature of the chain (default 1. MCMC only).
-
thin_factor: set it to produce a
file_root_thin.txt file containing every thin_factorth
sample (MCMC only. Usually, not needed with MultiNest).
-
thin_cool: cooling factor applied
in the thin process. It has to match
the temperature of the chain (default 1).
-
adjust priors: performs rough
importance sampling. Write your own
AdjustPriors routine.
-
map_params: set it to T to map
chain parameters to any function of the parameters
(e.g, transform from linear to log scale for plotting.
This will not adjust the prior,
though! use
adjust_priors instead).
Write your own MapParameters routine.
-
contour1, contour2: percentage of
confidence levels contours.
-
force_twotail: set it to T to force
2-tails limits on all variables regardless of
the settings for tailsxxx below.
-
plotparams_num: how many variables
to get plots for. If zero, uses all parameters which
have labels in .info file plus all added parameters
(with labels labAxxx).
This setting can be exceedingly slow, so use with care.
-
plotparams: list of parameter
numbers to plot, must match
plotparams_num above. For the
parameters saved in the chain, look at the .info
file to determine which number corresponds to which
parameter.
For added parameters, use the syntax
A1, A2, ...,
where a capital A denotes that the number refers to
an added parameter (numbering for added parameters
goes from 1 up to the maximum determined by the
value in add_columns).
plot_1D_pdf: set to T to plot
the 1D Bayesian posterior pdf.
plot_1D_meanlike: set to T to plot
the 1D mean likelihood (mean is taken over the posterior). Notice this is only plotted in the 1D SM files (not in the 1D Matlab).
-
plot_1D_meanchisq: set to T to plot
the 1D mean chi-square. Notice this is only plotted in the 1D SM files (not in the 1D Matlab).
-
plot_1D_profile: set to T to plot
the 1D profile likelihood (both with SM and with Matlab)
-
plot_1D_likelihood:
set to T to plot
the 1D likelihood from the data when plotting the
observables
(such as the DM relic abundance, g-2, etc). The codes
determines automatically which variables have an associated
likelihood function and
takes the values from the likedata.f90 file.
-
plot_2D_param: set it to 0 to
produce 2D plots only of a list of parameters
combinations (specified below). Set it to the number
corresponding to the parameter you want to have 2D
plots of (plotted against all other
parameters that have labels).
-
plot_2D_num: number of parameters
combinations for which to produce a 2D plot
(mean quality of fit and marginal probability density
will be both plotted by default and saved in different
.ps files). Only relevant if plot_2D_param
= 0. You must then specify a list of
plot1, plot2, ... giving the couples of
parameters that you want to plot against each other.
Use A to denote added variables, same syntax as above.
-
plot_contours: set to T to plot contours
in 2D plots. Contours will be plotted for the posterior
pdf and the profile likelihood, following the levels for
the 2 statistics.
-
plot_mean: set to T to plot the
posterior mean in the 1D (as a vertical bar)
and 2D plots (as a solid dot).
-
plot_bestfit: set to T to plot the best
fit value in the 1D (as a cross) and 2D plots
(as a cross with a circle around it).
-
plot_reference: set to T to plot a reference point of your choosing (hardcoded in the subroutine DefineRefPoint in mcsamples.f90. Plotted as a diamond.
-
plot_2D_meanlike: set to T to plot the mean likelihood in 2D.
-
plot_2D_meanchisq: set to T to plot the average chi-square in 2D.
-
color_scheme: color_scheme type for 2D plots. Options are: 1 for a smooth, continous colorscheme (default) or 2 for a discrete colour scheme with solid colouring of the contours.
-
all_2D_plots: set it to T if you
want to produce plotting data files for all possible
2D combinations (although the ones that will be
included in the .m file are still controlled by the
plot_2D_num variable above). This is
useful if you want to plot some other parameter
combination subsequently without having to re-run
GetPlots to get the corresponding matlab file.
Careful, setting this to T can produce several
thousands of plot files.
- colorbar_on: set to T to add a
colorbar to the bottom of 2D files.
-
num_3D_plots: number of 3D scatter
plots to produce. You must then specify a list of
3D_plot1, 3D_plot2, ... giving the
triplets of parameters that you want to plot
(x axis, y axis and coloured variable).
Use A to denote added variables, same syntax as above.
-
do_3D_plots: set it to T to produce
a single samples file used by 3D plots.
Setting is overriden to T if
num_3D_plots > 0.
-
colormap_name: put here the name of the colormap you want to use for 3D plots. see colormaps/ directory. If empty, uses default 'jet' colormap.
-
labA1, labA2: labels for added
parameters.
Parameters saved in the chain automatically get their
labels from the .info file.
-
limitsxx: lower and upper limit
for the 1D binning. Samples below/above the limits are
put in the first/last bin.
Use limitsA1, limitsA2, ...
for added variables instead. If you only want to
adjust the limits of the 1D plot, use
plot_limits instead.
-
plot_limitsxx: lower and upper limit
in the 1D and 2D plots of the corresponding variable
(purely cosmetic, does not affect the statistics).
-
tailsxxx: set to 1 for 1-tail
probabilities and to 2 for 2-tails (default).
For added variables use
tailsA1, tailsA2, ... instead.
Overridden if force_twotail = T.
-
cov_matrix_dimension: number of
parameters to get covariance matrix for. If you are
going to use the output as a proposal density make
sure you have map_params= F, and
the dimension equal to the number of
MCMC parameters of the run (9 for the CMSSM).
(top)
List of output
files and their content
-
Output files from MCMC runs:
those files are in the outroot directory.
-
.txt MCMC chains
-
.log log file containing details
of acceptance ratio and useful timing information if
timing=.true. in
source/paramdef.f90)
-
.info
a file containing the name and position of the saved
variables and other details of the run
-
Output files from MultiNest runs:
-
.txt the chains (posterior samples) produced by MultiNest. This file is updated after every 100 iterations.
-
equal_weights.dat updated after every 1000 iterations & contains the equally weighted posterior samples.
-
phys_live.points the current set of live points. This file is updated after every 100 iterations.
-
stats.dat contains the Bayesian evidence, mean, max likelihood & MAP parameters. If multimodal = T, it will contain local evidence values and mean, max
likelihood & MAP parameters for each found mode separately. This file is update after every 1000 iterations
-
resume.dat contains the information about check-pointing.
-
post_separate.dat is created only created if multimodal = T. It contains the chains (posterior samples) for modes with local log-evidence value, separated by 2
blank lines. Format is the same as .txt file.
-
Output files from getplots:
-
.margestats
marginalized 1D statistics for the Bayesian posterior.
Posterior mean point.
-
.likestats
1D statistics for the mean quality of fit.
Best fit point.
-
.proflstats 1D statistics
for the profile likelihood.
-
_1D.sm generates 1D plots
with SM.
-
_1D.m generates 1D plots
with matlab.
-
_2D.m generates 2D plots
with matlab.
-
_3D.m generates 3D plots
with matlab.
-
.burnin.sm generates a
burn-in plot for MCMC chains with SM (useful for
MCMC diagnostics).
-
.chain_loc.sm generates
with SM a plot showing the location of the chain
in parameter space as a function of step number
(useful for MCMC diagnostics).
(top)
Known issues
An up-to-date list of known issues with the current version (including patches and solutions when available) is maintained here. Please do feel free to contribute your own bug reports and suggestions for fixes.
|
 The graph and the table below show the scaling of the performance of SuperBayeS with the number of CPUs used. All runs have been performed on 2.8 GHz, 1600 MHz FSB Intel processors using
The graph and the table below show the scaling of the performance of SuperBayeS with the number of CPUs used. All runs have been performed on 2.8 GHz, 1600 MHz FSB Intel processors using